Single Pass DNA Sequencing
A regular but robust DNA sequencing reaction encompasses the cycle sequencing reactions, dye terminator removal and subsequent analysis on our Genetic Analyzers.
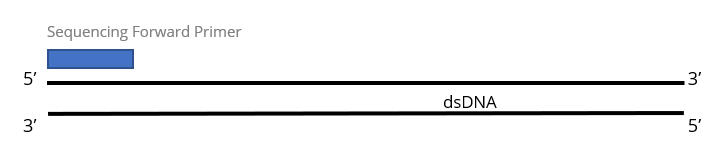
Purified DNA template is required for each sequencing primer and will be charged as 1 reaction. Most customers submit purified PCR products or purified plasmid DNA for this service.
1x Single Pass DNA Sequencing reaction:
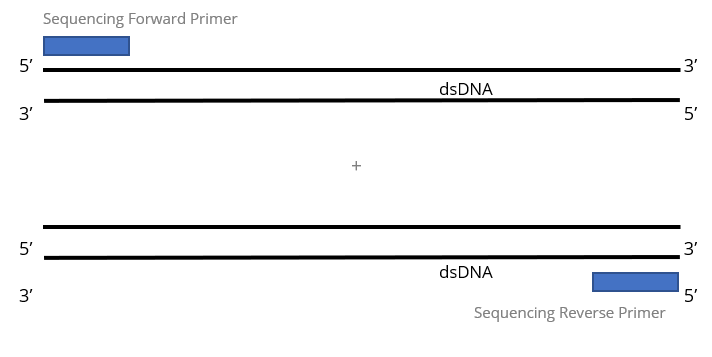
2x Single Pass DNA Sequencing reactions:
High Throughput (HT) DNA Sequencing
We also accept samples in 96-well plates for HT DNA sequencing. This is recommended for bulk orders, with a minimum of 80 reactions per plate. Unlike most sequencing company that only allow a few sequencing primers to be used from each 96-well plate, we have no limitation on the choices of sequencing primers for each plate. This is all thanks to our automation in liquid handling system that has no problem to pick-up any DNA template or sequencing primers at any of the combination that you would possibly order.
Difficult Template Sequencing
Difficult Template Sequencing (DTS) caters specifically to DNA templates with sequence context difficulties or large size. Sequence context difficulties include high G%, high C%, high GC%, repeat regions, secondary structure or loop structures found in shRNA vectors.
1st BASE adopts a robust protocol that maximizes the success rate of sequencing through these regions. However, the success rates of GT and GC repeats have shown to be limited by this chemistry. Currently, there is no guaranteed to overcome any homopolymeric regions. Please refer our troubleshooting guide for more details.
DTS also works for large DNA templates like purified genome DNA or purified BAC cloned DNA. If the same pair of primers able to amplify the target region under regular PCR condition, the same PCR primer will work well for DTS in individual sequencing reaction. But if you find the PCR primers hardly to amplify any fragment from such a large DNA template, we suggest you don’t try your luck with Sanger sequencing that is less forgiven for weak priming site. You may change your approach using Next-Generation Sequencing to sequence the large DNA template directly.
If you are not sure whether your current working of DNA template is difficult template for DNA sequencing, you most likely ordering our robust Single Pass DNA Sequencing services. Within a week after you receive the results, if there is sufficient remaining DNA template and sequencing primer for us to work on, you may request us to repeat the failed reaction using our difficult template protocol to evaluate the effectiveness of the chemistry. The troubleshooting of difficult template sequencing is always free of charge. We like the challenges and thus please send us more.
M13-Tailed Sequencing
M13-tailed sequencing has been long available since manual slab gel electrophoresis on DNA Sequencer. In most of the disease screening that requires Sanger DNA sequencing as confirmatory method, scientists add the M13 primer sequences to be adjacent with their gene specific primers. So that the generated PCR products will be extended with additional M13 primer sequences. With this, all sequencing confirmatory will use the same M13 sequencing primer in sequencing reaction. And since most of the sequencing company provides M13 sequencing primers as free, you don’t even need to send your sequencing primer in each order; hassle-free.
In most of the viral sequence amplification, its PCR/ qPCR primers typically has degenerated bases to increase its viability to amplify as many variants as possible. For Sanger sequencing reaction that is less forgiven for weak priming efficiency, you may experience inconsistent results due to the presence of the degenerate bases because most of the project owner will use the PCR/ qPCR primers as their sequencing primer. M13-tailed sequencing hence beautifully addressed these problems to change its sequencing primer to M13 primers as default.
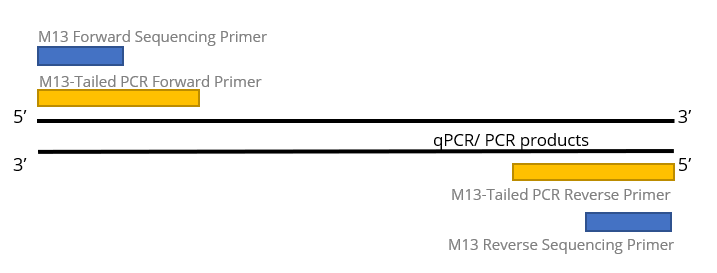
1st BASE M13-tailed sequencing includes quantification of DNA templates, M13-tail PCR, PCR product purification, bi-directional sequencing, and alignment to reference sequence. Customer to provide (i) unpurified qPCR/ PCR products, (ii) qPCR/ PCR primer sequences, and (iii) reference sequence for each product to tabulate the variant/ mutation.
For ready COVID-19 qPCR products, we offer free PCR optimization before M13-tailed sequencing for some listed genes. For other gene of disease screening, a PCR optimization charge will be applied only for the first order. For subsequence sequencing order, only M13-tailed sequencing fee will be charge for each DNA template.
For customers that already had their purified M13-tailed qPCR/ PCR products, they can order Single Pass DNA Sequencing (SS1001) directly. We provide wide range of M13 sequencing primers for free. Please check the availability of your chosen M13 primer sequences, then you don’t even need to send us your sequencing primer in each new sequencing order.
*** Available to Malaysia & International Customers only.
Ready-to-Load Sequencing
For Ready-to-Load samples, customers undertake the first two steps in Automated Sanger Sequencing, which are (i) cycle sequencing reaction and (ii) post-sequencing clean-up; then only send us the dried cycle sequencing products. We will dissolve the dried cycle sequencing products, denature, and load into our Genetic Analyzer for electrophoresis and automated base-calling.
We accept reactions performed with ABI® BigDye Terminator Cycle Sequencing Kit v3.1. If you are using other sequencing chemistry kit, please acquire before we confirm its acceptance to load into our Genetic Analyzers.
About your setup of cycle sequencing reaction
Many of our Ready-to-load customers have modified or optimised their sequencing reactions with a novel protocol, for example diluting BigDye® sequencing mix, using an increased number of cycles, using additive like DMSO and etc. We may not be able to provide support for these modified reactions, but we could assure you that our Genetic Analyzers are maintained in the tip-top condition. A control reaction will be run together with your ready-to-load reaction to certify our electrophoresis conditions are always passing our QC benchmark.
About your post-cycle sequencing clean-up
We recommend fresh cycle sequencing reactions to be purified immediately right off from its thermocycler. If this cannot be done, take them out from your thermocycler and freeze them at -20°C over the weekend before post sequencing clean-up.
You may use the standard EDTA-Sodium Acetate-Ethanol Precipitation (which is recommended by Applied Biosystems for BigDye® Terminator Cycle Sequencing Kit v3.1). This requires skilful handling. Other clean-up methods in the market for post-sequencing include column/ plate purification using gel bed filtration, magnetic beads, ethanol precipitation with co-precipitant etc. If there is no carry-over of ethanol residue or water after the drying process, your samples should be fine and Ready-to-Load. For reaction(s) purified by magnetic beads, please indicate the elution volume (by water).
You are not required to add deionised formamide or Hi-Di® formamide on your Ready-to-Load samples. We will add formamide and perform the denaturation on our thermocyclers prior loading.
Ready-to-Load DNA Sequencing with Clean Up
Customer may send us their cycle sequencing reactions in aluminium wrapped single tubes or 96-well plate. We will perform the post sequencing clean-up to remove excess dye terminators and the subsequent analysis on our Genetic Analyzers.
We accept reactions performed with ABI® BigDye Terminator Cycle Sequencing Kit v3.1. If you are using other sequencing chemistry kit, please acquire before we confirm its acceptance to load into our Genetic Analyzers.
Both cycle sequencing reaction setup and post-cycle sequencing clean-up are equally crucial to high quality of DNA sequencing results. Poor handling in either step will compromise the result significantly.
We would love to help in any troubleshooting of customers’ cycle sequencing reaction setup and post sequencing clean-up. Please contact your Local Sales Office for assistance. Charges may apply. You can send us the following: -
- The DNA template and its sequencing primers in separate tubes (as in our service SS1001);
- Cycle sequencing reactions, which is without post sequencing clean-up (as in our service SS1019); and
- Ready-to-Load samples, where the customer performs the cycle sequencing reactions and post sequencing clean-up at their own (as in our service SS1005).
Contact Us for Orders or Enquiries
Select Your Country
The following is the list of complimentary Universal Primers offered by our DNA Sequencing facility.
| Universal Primers | Sequence (5' - 3') |
| 1492R | 5' TACGGYTACCTTGTTACGACTT 3' |
| 27F | 5' AGAGTTTGATCMTGGCTCAG 3' |
| 35S-A | 5' AAGGGTCTTGCGAAGGATAG 3' |
| 35S-B | 5' AGTGGAAAAGGAAGGTGGCT 3' |
| 518F | 5' CCAGCAGCCGCGGTAATACG 3' |
| 800R | 5' TACCAGGGTATCTAATCC 3' |
| AD Reverse | 5' AGATGGTGCACGATGCACAG 3' |
| a-Factor | 5' TACTATTGCCAGCATTGCTGC 3' |
| AOX1 Forward | 5' GACTGGTTCCAATTGACAAGC 3' |
| AOX1 Reverse | 5' GCAAATGGCATTCTGACATCC 3' |
| BGH-R | 5' TAGAAGGCACAGTCGAGG 3' |
| Bluescript KS | 5' TCGAGGTCGACGGTATC 3' |
| Bluescript SK | 5' CGCTCTAGAACTAGTGGATC 3' |
| CMV-F | 5' CGCAAATGGGCGGTAGGCGTG 3' |
| CMV-profor | 5' ATGGGCGGTAGGCGTG 3' |
| CYC1 Reverse | 5' GCGTGAATGTAAGCGTGAC 3' |
| DsRed1-C | 5' AGCTGGACATCACCTCCCACAACG 3' |
| DsRed1-N | 5' GTACTGGAACTGGGGGGACAG 3' |
| EBV-RP | 5' GTGGTTTGTCCAAACTCATC 3' |
| EGFP-C | 5' CATGGTCCTGCTGGAGTTCGTG 3' |
| EGFP-CF | 5' AGCACCCAGTCCGCCCTGAGC 3' |
| EGFP-CR | 5' CGTCCATGCCGAGAGTG 3' |
| EGFP-N | 5' CGTCGCCGTCCAGCTCGACCAG 3' |
| EGFP-NR | 5' CGTCGCCGTCCAGCTC 3' |
| GAL1 Forward | 5' AATATACCTCTATACTTTAACGTC 3' |
| Gal4AD | 5' TACCACTACAATGGATG 3' |
| GLprimer1 | 5' TGTATCTTATGGTACTGTAACTG 3' |
| GLprimer2 | 5' CTTTATGTTTTTGGCGTCTTCCA 3' |
| HCO2198 | 5' TAAACTTCAGGGTGACCAAAAAATCA 3' |
| hU6-F Primer | 5’-GAGGGCCTATTTCCCATGATT-3’ |
| ITS1 | 5' TCCGTAGGTGAACCTGCGG 3' |
| ITS2 | 5' GCTGCGTTCTTCATCGATGC 3' |
| ITS3 | 5' GCATCGATGAAGAACGCAGC 3' |
| ITS4 | 5' TCCTCCGCTTATTGATATGC 3' |
| ITS5 | 5' GGAAGTAAAAGTCGTAACAAGG 3' |
| KAN2-FP | 5' ACCTACAACAAAGCTCTCATCAACC 3' |
| KAN2-RP | 5' GCAATGTAACATCAGAGATTTTGAG 3' |
| LCO1490 | 5' GGTCAACAAATCATAAAGATATTGG 3' |
| LpJET1.2F | 5' CTGCTTTAACACTTGTGCCTGA 3' |
| LpJET1.2R | 5' TTCCTGATGAGGTGGTTAGCAT 3' |
| M13F (-20) | 5' GTAAAACGACGGCCAGT 3' |
| M13F (-29) | 5' CACGACGTTGTAAAACGAC 3' |
| M13-FP | 5' TGTAAAACGACGGCCAGT 3' |
| M13F-pUC(-40) | 5' GTTTTCCCAGTCACGAC 3' |
| M13R (-20) | 5' GCGGATAACAATTTCACACAGG 3' |
| M13R (-24) | 5' GGAAACAGCTATGACCATG 3' |
| M13R-pUC (-26) | 5' CAGGAAACAGCTATGAC 3' |
| MT Forward | 5' CATCTCAGTGCAACTAAA 3' |
| pBacPAC-RP | 5' GTCTGTAAATCAACAACGC 3' |
| pBAD-F | 5' ATGCCATAGCATTTTTATCCA 3' |
| pBAD-FP | 5' ATGCCATAGCATTTTTATCC 3' |
| pBAD-R | 5' GATTTAATCTGTATCAGG 3' |
| pBRrevBam | 5' GGTGATGTCGGCGATATAGG 3' |
| pDONOR-FP | 5' TAACGCTAGCATGGATCTC 3' |
| pEGFP_N | 5' CCGTCCAGCTCGACCAG 3' |
| pEGFP-FP | 5' TTTAGTGAACCGTCAGATC 3' |
| pEGFP-RP | 5' AACAGCTCCTCGCCCTTG 3' |
| pESP-RP | 5' TCCAAAAGAAGTCGAGTGG 3' |
| pET-24a | 5' GGGTTATGCTAGTTATTGCTCAG 3' |
| pET-RP | 5' CTAGTTATTGCTCAGCGG 3' |
| pFastBac Forward | 5' GGATTATTCATACCGTCCCA 3' |
| pFastBac Reverse | 5' CAAATGTGGTATGGCTGATT 3' |
| pGEX3 | 5' GGAGCTGCATGTGTCAGAGG 3' |
| pGEX5 | 5' GGCAAGCCACGTTTGGTG 3' |
| pJET1.2F | 5' CGACTCACTATAGGGAGAGCGGC 3' |
| pJET1.2R | 5' AAGAACATCGATTTTCCATGGCAG 3' |
| pMalE | 5' TCAGACTGTCGATGAAGC 3' |
| pQE-F | 5' CCCGAAAAGTGCCACCTG 3' |
| pQE-R | 5' GTTCTGAGGTCATTACTGG 3' |
| pREP-fwd | 5' GCTCGATACAATAAACGCC 3' |
| pRH Forward | 5' CTGTCTCTATACTCCCCTATAG 3' |
| pRH Reverse | 5' CAAAATTCAATAGTTACTATCGC 3' |
| pTrcHis Forward | 5' GAGGTATATATTAATGTATCG 3' |
| QE Promoter | 5' CCGAAAAGTGCCACCTG 3' |
| RVprimer3 | 5' CTAGCAAAATAGGCTGTCCC 3' |
| RVprimer4 | 5' GACGATAGTCATGCCCCGCG 3' |
| SP6 | 5' ATTTAGGTGACACTATAG 3' |
| STag 18mer Primer | 5' GAACGCCAGCACATGGAC 3' |
| SV40-pArev | 5' CCTCTACAAATGTGGTATGG 3' |
| SV40-Promoter | 5' GCCCCTAACTCCGCCCATCC 3' |
| T3 | 5' ATTAACCCTCACTAAAG 3' |
| T7 | 5' AATACGACTCACTATAG 3' |
| T7 EEV | 5' ATGTCGTAATAACCCCGCCCCG 3' |
| T7promoter | 5' TAATACGACTCACTATAGGG 3' |
| T7terminator | 5' GCTAGTTATTGCTCAGCGG 3' |
| U-19mer Primer | 5' GTTTTCCCAGTCACGACGT 3' |
| U6 Primer | 5’-GGGCAGGAAGAGGGCCTAT-3’ |
| SP6Long | 5'-ATTTAGGTGACACTATAGAATAC-3' |
| T7Long | 5'-GTAATACGACTCACTATAGGGC-3' |
| LpJet1_2F | 5' CTGCTTTAACACTTGTGCCTGA 3' |
| LpJet1_2R | 5' TTCCTGATGAGGTGGTTAGCAT 3' |
| 785F | 5' GGATTAGATACCCTGGTA 3' |
| 907R | 5' CCGTCAATTCMTTTRAGTTT 3' |
Guides on Preparations and Requirements
![]() Sample Requirements
Sample Requirements![]() List of Services & Details
List of Services & Details![]() Guidelines to Sample Preparation and Quantification
Guidelines to Sample Preparation and Quantification
